CRISPR maps temperature-sensitive mutations in E. coli
CRISPR maps temperature-sensitive mutations in E. coli

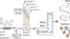
The Link Lab has published a study in which they used the CRISPR technology to explore the consequences of single amino acid changes in E. coli. They utilized CRISPR's fast and versatile gene-editing ability to construct over 15,000 E. coli mutants. What makes these mutants unique? Each mutant has a single amino acid change in one essential protein. More than a thousand of these mutants showed temperature sensitivity, which means that they grow faster at 30°C than at 42°C. Following up on the temperature-sensitive mutants, the researchers found distinct changes in metabolism when these bacteria were stopped from growing at 42°C. Some mutants accumulated metabolites others decoupled their metabolism from growth. For example, a mutant modified in its homoserine kinase gene, displayed long-lasting homoserine production, with production rates being tunable by temperature variations. Another discovery centered on a DNA polymerase mutant, offering new technologies in modulating E. coli growth and overproduction of chemicals.
"The research paves a new pathway in our understanding of amino acid changes and their functional impacts," says Hannes Link. "Harnessing CRISPR's power to map the functional consequences of single amino acid changes could redefine boundaries in biotechnology and medical research." The vision of the Link Lab is to use the rich data from such CRISPR screens for future applications in machine learning. "The scale of data we've gathered has the potential to be a great training source for machine learning models, and predicting not only protein structure but also function from sequence data" Link adds.
The study was published in Molecular Systems Biology, and is featured on the cover of the October issue.
Publication:
Schramm T, Lubrano P, Pahl V, Stadelmann A, Verhülsdonk A, Link H. Mapping temperature-sensitive mutations at a genome scale to engineer growth switches in Escherichia coli. Mol. Syst. Biol. (2023) doi:
10.15252/msb.202311596.
For further information, find the paper here.
Read this comment about the paper.
Prof. Dr. Hannes Link
Professor for Bacterial Metabolomics
Leon Kokkoliadis
Public Relations Management
Tel: +49 7071 29-74707
E-Mail: leon.kokkoliadis@uni-tuebingen.de
Related Articles
Springer Nature



Springer Nature


